Document Type
Dataset
Publication Date
6-2019
Digital Object Identifier
https://doi.org/10.35482/bld.001.2019
Data Directories
[ Wild Type, You-too] | -- Raw ……| -- AT ………..| -- *.h5 ………..| -- ilastik.ilp .…..| -- Gfap ………..| -- *.h5 ………..| -- ilastik.ilp | -- Probability ……| -- AT ………...| -- *_Probabilities.h5 ……| -- Gfap ………...| -- *_Probabilities.h5 | -- PSI ……| -- AT_*.psi ……| -- Gfap_*.psi ……| -- model.csv | -- landmarks.csv | -- PSI-Independent alignment check ……| -- AT_*.psi ……| -- Gfap_*.psi ……| -- model.csv | -- landmarks_align.csv
Experiment Description
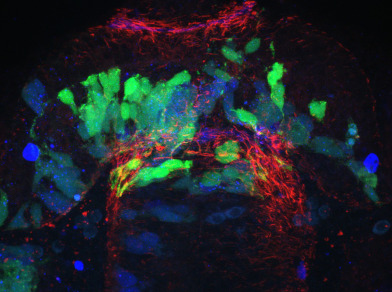
The published Gli2DR (you-too, yot) exhibits significant and obvious commissural defects in the zebrafish diencephalic post-optic commissure (POC). The POC is a parabolic ribbon like structure which connects the two halves of the ventral anterior nervous system. It develops in concert with a host of glial cells, called a glial bridge, which has been proposed by us and others to perform the region for POC development. We have built ΔSCOPE as a computational analytical tool to quantify POC and glial bridge development. We use the visually apparent defects in POC development to ascertain whether ΔSCOPE is able to detect POC defects and whether they agree with the visual appearance of the you-too mutant. We have run ΔSCOPE alignment on wild type and you-too POCs twice with independent user input to determine whether results are reproducible. We have compared these results with the original semi-quantitative analysis published in Barresi et al. 2005. We find that our results are in agreements with these findings. We also observe that ΔSCOPE exhibits greater sensitivity to the position and axis of POC disturbances than in Barresi 2005, as well as greater statistical power.
Data Description
Raw: After collection on a Leica LSCM, data from each sample and each channel were saved individually to hdf5 files. The files are saved in a subdirectory for each channel as appropriate. Additionally, the ilastik file that was used to process the raw data is included in the subdirectory for each channel. Probability: Raw data files were processed using the ilastik training file saved in each raw channel subdirectory. The output from ilastik is a two channel hdf5 file saved to the appropriate channel subdirectory. PSI: After processing through ΔSCOPE, the resulting data is saved to a single directory as a psi file. The model file contains the coefficients that specified the quadratic function fit to each sample.
Collection Method
28 hpf zebrafish embryos from wild type and you-too (gli2DR) homozygote mutant backgrounds (you-too embryos were screened for mutant phenotypic markers for selection) were fixed in 4% PFA for 3 hrs (or o/n 4ºC). Immunocytochemistry using mouse IgG2b anti-acetylated tubulin and mouse IgG1 anti-ZRF1 (ZIRC) (anti-Gfap) made up in PBStx (2% triton x100) with 5% BSA and 10% normal goat serum. Secondaries used were alexafluor goat anti-mouse IgG2b 488 and goat anti-mouse IgG1 594 (invitrogen). Heads were dissected and mounted for coronal collection of z-stacks imaging from most anterior of the POC to the the start of the ventral rostral cluster. Images were collected on a leica scanning confocal microscope (LSCM) SP5, using an argon 488 laser and a 594 laser. Care is taken to ensure that dichroic mirrors do not pick up signal from adjacent channels. Imaging is performed at 63x with a 1.5x digital zoom with a pixel setting of 1024 x 1024, yielding a 0.128 x 0.128 um xy dimension, and a z thickness of 0.21 um is selected. Post collection, images are imported into Fiji/Imagej, and collections are cropped to select only the immediate surrounding of the POC, approximately from the luminal junction of the telencephalon and diencephalon to a corresponding distance in the ventral direction on the other side of the POC. Care is taken to eliminate any AT signal contributions of the telencephalic anterior commissure. After cropping, any associated channels are then split, and saved separately as HDF5 files for Ilastik processing.
File Formats
HDF5: Image-based data are saved as hdf5 files, which are compatible with Ilastik. The file format is described here https://portal.hdfgroup.org/display/support. Hdf5 files can be easily viewed using the plugin for Fiji https://github.com/fiji/HDF5_Vibez/. ILP: These files are generated by the open source machine learning program ilastik https://github.com/fiji/HDF5_Vibez/. They contain the training data used to generate the probability files. PSI: Text based file format similar to a csv with space delimiters. The first twenty lines of the file specify details about the file including the columns: x, y, z, alpha, theta, r.
Recommended Citation
Barresi Lab, Smith College, "Validation Experiment: Wild Type, Raw, AT" (2019). Dataset, Smith College, Northampton, MA.
https://scholarworks.smith.edu/bl_ds_yte/3


Comments
This is file contains the zipped AT folder from the Raw folder
All files in the Validation Experiment include the experiment's .json and .csv files as additional files.