ΔSCOPE DATA REPOSITORY
Data sets in collection are associated with the forthcoming paper:
ΔSCOPE: A new method to quantify 3D biological structures and identify differences in zebrafish forebrain development
Link to pre-print hereAbstract: Research in developmental biology has traditionally relied on the analysis of clear morphological phenotypes. Increasingly, this data is collected using high spatial resolution image capture techniques. Despite these advances in resolution, maximum intensity projections (MIPs), which spatially compress this data, have continued to be the standard practice for visualization.
However, as biology turns towards the analysis of more subtle phenotypes, MIPs and qualitative analysis are reaching the end of their useful life, and a substantial need for new quantitative approaches is apparent. Recent advances in computational power now enable analysis of the 3D spatial relationships and signal distribution contained in 3D data sets.
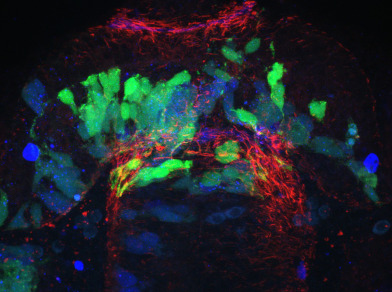
We have developed a program ∆SCOPE, which enables quantification and statistical analysis of spatial relationships and signal density in 3D multichannel signals that are positioned around a well-defined structure contained in a reference channel. We have validated the power of ∆SCOPE analysis by analyzing and quantifying normal axon and glial cell guidance in the zebrafish forebrain as well as testing it on previously published phenotypes resulting from abnormal Slit guidance cues in the zebrafish forebrain. ∆SCOPE is able to detect subtle previously uncharacterized changes in zebrafish forebrain midline crossing axons and glia, and can do this despite disruption to the primary structural channel. We propose that ∆SCOPE is an innovative approach to advancing the state of image quantification in the field of high resolution microscopy, and that the techniques presented here are of broad use to the field. This method is developed as a user-friendly, open source program, which requires some computer skills, that the community at large can use to quantify 3D, multichannel datasets, which will provide statistical rigor and novel insight often lost in the qualitative inspection of subtle phenotypic changes.
DeltaScope DOI: https://doi.org/10.35482/bld.06.2019